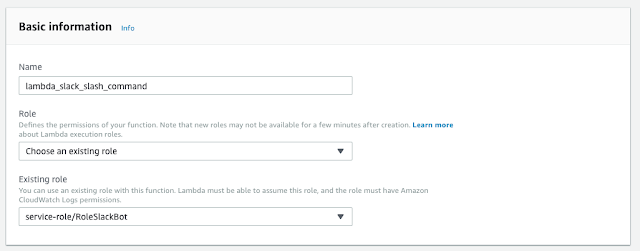
Install SAM CLI
$ sudo pip install --user --upgrade aws-sam-cli
Get a list of existing Lambda functions
aws lambda list-functions
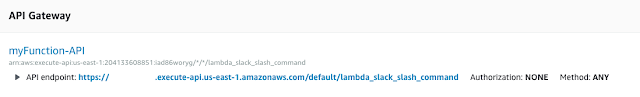
Using Postman to test AWS API Gateway
- download Postman app
$ sudo pip install --user --upgrade aws-sam-cli
aws lambda list-functions
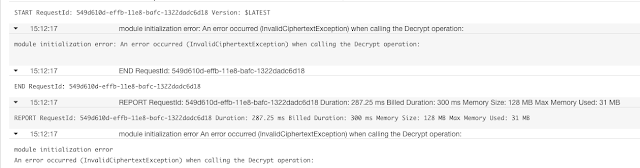
$ pip install jupyter_contrib_nbextensions
$ jupyter contrib nbextension install --user
$ jupyter notebook
 Detroit Autonomous Vehicle and Robotics Meetup
Detroit Autonomous Vehicle and Robotics Meetup  The new LattePanda Alpha Single Board Computer has ample 8GB of RAM, operates on 12V, can run a MacOS with TuriCreate Machine Learning and use massive GPU connected via the M.2 bus, hence making it an excellent in-vehicle machine learning platform.
The new LattePanda Alpha Single Board Computer has ample 8GB of RAM, operates on 12V, can run a MacOS with TuriCreate Machine Learning and use massive GPU connected via the M.2 bus, hence making it an excellent in-vehicle machine learning platform. References:
https://arxiv.org/abs/1810.04719
https://github.com/google/uis-rnn
https://catalog.ldc.upenn.edu/LDC2001S97
| Raspberry Pi 3B | 7-inch touchscreen | 7-inch case | 10.1 touchscreen | Infrared camera |
$ diskutil list
/dev/disk2 (external, physical):
#: TYPE NAME SIZE IDENTIFIER
0: FDisk_partition_scheme *15.9 GB disk2
1: Windows_FAT_32 PI3_RASBIAN 15.9 GB disk2s1
$ sudo diskutil unmountDisk /dev/disk2
Password:
Unmount of all volumes on disk2 was successful
$ sudo dd bs=1m if=/Users/uki/Downloads/2018-10-09-raspbian-stretch.img of=/dev/disk23944+0 records in3944+0 records out4135583744 bytes transferred in 3019.441172 secs (1369652 bytes/sec)
$ sudo diskutil eject /dev/disk2Password:Disk /dev/disk2 ejected
# shorthand
# sframe[3]['image'].show()
# image_testing_SFrame[0:5]['image'].explore()
sframe = image_testing_SFrame[0:5] # show first 5 records
def show_images(sframe, image_column="image", label_column="label"):
for subset_dictionary in sframe:
image = subset_dictionary[image_column]print(subset_dictionary[label_column])
image.show()
show_images(sframe, image_column="image", label_column="label")
show_images(sframe)
dogs_SFrame = image_training_SFrame.filter_by(values="dog", column_name="label", exclude=False)
print(dogs_SFrame["label"][0:15])
['dog', 'dog', 'dog', 'dog', 'dog', 'dog', 'dog', 'dog', 'dog', 'dog', 'dog', 'dog', 'dog', 'dog', 'dog']
animals = ["dog", "cat", "bird"]
animals_SFrame = image_training_SFrame.filter_by(values=animals, column_name='label', exclude=False)
print(animals_SFrame["label"][0:15])
['bird', 'cat', 'cat', 'dog', 'bird', 'dog', 'bird', 'bird', 'cat', 'dog', 'cat', 'bird', 'cat', 'cat', 'dog']
training_sframes = {}
for label in unique_labels:
#print (label)
training_sframes[label] = image_training_SFrame.filter_by(
values = label,
column_name = "label",
exclude = False)
for key_name in training_sframes: # dictionary training_sframes
print(key_name)
labels_column_SArray = SFrame_DataSet['label']
print(type(labels_column_SArray))
unique_labels = labels_column_SArray.unique()
print(unique_labels)
['bird', 'dog', 'cat', 'automobile']
 I have learned that none of my GUI Mac programs were able to expand the 13 GB dataset, however, the command line had no problem with it.
I have learned that none of my GUI Mac programs were able to expand the 13 GB dataset, however, the command line had no problem with it.$ tar xvzf BIG_DATASET_MANY_THOUSANDS_FOLDERS.tar.gz
$ tar xvfz BIG_DATASET_MANY_THOUSANDS_FOLDERS.tar.gz /directory_path
# parameters:
# -a --archive; look at everything recursively
# -i; --itemize-changes; print update about each file
# -h; --human-readable
# -W; --whole-file; avoid file deltas
# --progress; show progress in terminal
# --log-file=XYZ.log; log the progress to file, this might be useful when resuming
$ rsync -aW source_directory/ destination_directory/
$ conda env export > environment_turi_20181105.yml
rm -r .... /anacondaX/
conda env create -f environment_turi_20181105.yml
$ conda activate turi
$ conda env list
# conda environments:
#
/Users/uki/.julia/conda/3
/Users/uki/.julia/packages/ORCA/uEiWT/deps
base /anaconda2
turi * /anaconda2/envs/turi
python -m ipykernel install --user --name turi --display-name "Python 2.7 (turi)"
$ jupyter notebook
$ source activate py36
$ conda env list
# conda environments:
/Users/uki/.julia/conda/3
/Users/uki/.julia/packages/ORCA/uEiWT/deps
base /Volumes/DATA/anaconda3
py2 /Volumes/DATA/anaconda3/envs/py2
py36 * /Volumes/DATA/anaconda3/envs/py36
$ conda install -c derickl turicreate
$ conda install ipykernel
$ conda update --all
python -m ipykernel install --user --name py36 --display-name "Python 3.6 Turi (env py36)"
Installed kernelspec py36 in /Users/uki/Library/Jupyter/kernels/py36
$ conda env export > environment_py36_20181102.yml
$ jupyter notebook
import turicreate as turi
WARNING: You are using MXNet 1.2.1 which may result in breaking behavior. To fix this, please install the currently recommended version: pip uninstall -y mxnet && pip install mxnet==1.1.0 If you want to use a CUDA GPU, then change 'mxnet' to 'mxnet-cu90' (adjust 'cu90' depending on your CUDA version):
(py36) $ pip uninstall -y mxnet && pip install mxnet==1.1.0
import Pkg;
Pkg.add("PyCall")
Updating registry at `~/.julia/registries/General` Updating git-repo `https://github.com/JuliaRegistries/General.git` [1mFetching: [========================================>] 100.0 %.0 %
Resolving package versions...
Updating `~/.julia/environments/v1.0/Project.toml` [5c1ef01f]
- Cookbook v0.3.0 [5befdd30] - Hilbert v0.1.0 [5c077215]
- LatticeSites v0.2.0 Updating `~/.julia/environments/v1.0/Manifest.toml` [5c1ef01f]
- Cookbook v0.3.0 [5befdd30]
- Hilbert v0.1.0 [5c077215]
- LatticeSites v0.2.0
Pkg.update()
Updating registry at `~/.julia/registries/General`
Updating git-repo `https://github.com/JuliaRegistries/General.git`
Resolving package versions...
Installed KernelDensity ── v0.5.1
Installed Interpolations ─ v0.9.2
Updating `~/.julia/environments/v1.0/Project.toml` [5cadff95]
- JuliennedArrays v0.1.0 [5ab0869b] ↑ KernelDensity v0.5.0 ⇒ v0.5.1 [5c958174]
- SuperEnum v0.1.3 Updating `~/.julia/environments/v1.0/Manifest.toml` [a98d9a8b] ↓ Interpolations v0.10.5 ⇒ v0.9.2 [5cadff95]
- JuliennedArrays v0.1.0 [5ab0869b] ↑
KernelDensity v0.5.0 ⇒ v0.5.1 [5c958174]
- SuperEnum v0.1.3
using PyCall
SYSTEM: show(lasterr) caused an error Stacktrace: [1] _include_from_serialized(::String, ::Array{Any,1}) at ./loading.jl:630 [2] macro expansion at ./logging.jl:312 [inlined] [3] _require_search_from_serialized(::Base.PkgId, ::String) at ./loading.jl:701 [4] _require(::Base.PkgId) at ./loading.jl:934 [5] require(::Base.PkgId) at ./loading.jl:855 [6] macro expansion at ./logging.jl:311 [inlined] [7] require(::Module, ::Symbol) at ./loading.jl:837 [8] top-level scope at In[1]:1The error above prevents the execution of the below:
KERNEL EXCEPTION
SYSTEM: show(lasterr) caused an errorStacktrace:
[1] #invokelatest#1 at ./essentials.jl:697 [inlined]
[2] invokelatest at ./essentials.jl:696 [inlined]
[3] eventloop(::ZMQ.Socket) at /Users/uki/.julia/packages/IJulia/0cLgR/src/eventloop.jl:8
[4] (::getfield(IJulia, Symbol("##12#15")))() at ./task.jl:259
@pyimport math
#math.sin(math.pi / 4) - sin(pi / 4) # returns 0.0
@pyimport time
@pyimport numpy.random as nr nr.rand(3,4)
$ jupyter notebook
Traceback (most recent call last):
File "/anaconda3/bin/jupyter-notebook", line 11, in
sys.exit(main())
File "/anaconda3/lib/python3.7/site-packages/jupyter_core/application.py", line 266, in launch_instance
return super(JupyterApp, cls).launch_instance(argv=argv, **kwargs)
File "/anaconda3/lib/python3.7/site-packages/traitlets/config/application.py", line 657, in launch_instance
app.initialize(argv)
File "", line 2, in initialize
File "/anaconda3/lib/python3.7/site-packages/traitlets/config/application.py", line 87, in catch_config_error
return method(app, *args, **kwargs)
File "/anaconda3/lib/python3.7/site-packages/notebook/notebookapp.py", line 1602, in initialize
self.init_webapp()
File "/anaconda3/lib/python3.7/site-packages/notebook/notebookapp.py", line 1381, in init_webapp
self.http_server.listen(port, self.ip)
File "/anaconda3/lib/python3.7/site-packages/tornado/tcpserver.py", line 143, in listen
sockets = bind_sockets(port, address=address)
File "/anaconda3/lib/python3.7/site-packages/tornado/netutil.py", line 168, in bind_sockets
sock.bind(sockaddr)
OSError: [Errno 49] Can't assign requested address
$ jupyter notebook --ip=127.0.0.1
$ lsof | grep 8888Skype 407 uki txt REG 1,5
$ lsof -nP | grep 8888
_ _ _(_)_ | Documentation: https://docs.julialang.org (_) | (_) (_) | _ _ _| |_ __ _ | Type "?" for help, "]?" for Pkg help. | | | | | | |/ _` | | | | |_| | | | (_| | | Version 1.0.1 (2018-09-29) _/ |\__'_|_|_|\__'_| | Official https://julialang.org/ release|__/ |
julia> println("Hello World,", "Julia!")Hello World,Julia!
git clone https://github.com/JuliaLang/julia.git
julia> using Pkg
julia> Pkg.add("IJulia")
$ git clone https://github.com/stevengj/NBInclude.jl.git julia_NBInclude
$ ping Google.comPING Google.com (74.125.138.102) 56(84) bytes of data.
$ ssh dummy@74.125.138.102ssh: connect to host 74.125.138.102 port 22: Connection refused
$ python --version
$ python3 --version
$ python3
wget https://3230d63b5fc54e62148e-c95ac804525aac4b6dba79b00b39d1d3.ssl.cf1.rackcdn.com/Anaconda-2.3.0-Linux-x86_64.sh-bash: .: Anaconda-2.3.0-Linux-x86_64.sh: cannot execute binary file
$ chmod +x Anaconda-2.3.0-Linux-x86_64.sh
$ . Anaconda-2.3.0-Linux-x86_64.sh
$ java
$ sudo apt install default-jre
$ sudo apt install openjdk-11-jre-headless
$ sudo apt install openjdk-8-jre-headless
$ sudo apt install openjdk-11-jre-headless
$ java --versionopenjdk 10.0.1 2018-04-17
/Volumes/DATA/anaconda3/envs/py27/lib/python2.7/site-packages/graphlab/canvas/server.pyc108 self.__server = tornado.httpserver.HTTPServer(self.__application, io_loop=self.__loop)TypeError: initialize() got an unexpected keyword argument 'io_loop'Fix Attempt 1:
$ conda update tornado
environment location: /Volumes/DATA/anaconda3/envs/py27
tornado: 5.1-py27h1de35cc_0 --> 5.1.1-py27h1de35cc_0
Fix is not successful
Fix Attempt 2:Reinstall Anaconda3
% conda update --all -y
>> command not found: conda
curl
numpy
matplotlib
jupyter_core
protobuf
sqlite
ipython
jupyterlab
jupyter
notebook
matplotlib
pip
pandas
pillow
scikit-learn
python-3.9
$ conda activate base$ conda update -n base -c defaults conda$ conda update --all -y
cd $REPO // the directory you want as a base of your project (e.g. in GitHub directory)
jupyter-lab
% python --version
Python 3.9.13
$ conda create -n py_39_tf python=3.9 tensorflow -y
$ conda info --envs
% conda info --envs
# conda environments:
#
base * /Users/uki/opt/anaconda3
py_39_tf /Users/uki/opt/anaconda3/envs/py_39_tf
% conda activate py_39_tf
(py_39_tf) uki ~ %
$ python -m ipykernel install --user --name py_36_tf --display-name "Python 3.6 (tensorflow)"
$ ls -alt ~/Library/Jupyter/kernels/
$ rm -r ~/Library/Jupyter/kernels/my_old_kernel_name